Note
Go to the end to download the full example code.
Multi-slice 2D registration#
By default mdreg performs 3D coregistration on 3D series, and 2D coregistration on 2D series. In case the 3D series encodes a collection of 2D slices with gaps in between (multi-slice data), this may not be appropriate.
This examples shows how mdreg can be used to perform 2D coregistration slice-by-slice on a 3D series by setting the keyword argument force_2d=True.
Setup#
import numpy as np
import mdreg
Fetch the multi-slice MOLLI dataset
Perform slice-by-slice motion correction#
Since coregistration is performed in 2D, the deformation field only has two components:
print(f'The deformation field has {defo.shape[-1]} components.')
The deformation field has 2 components.
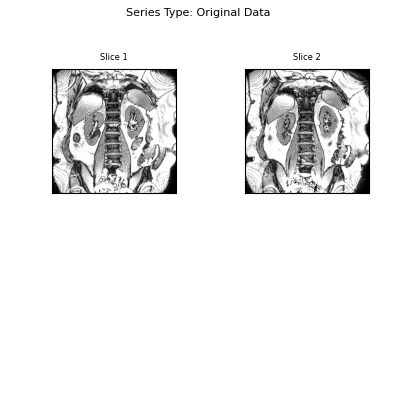
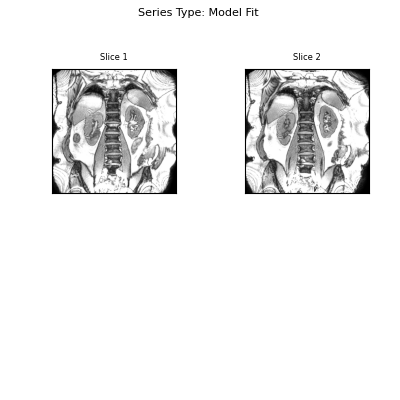
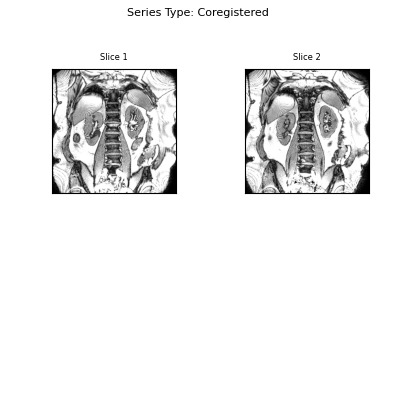
Visualise the results
Different options per slice#
It is not uncommon that each slice in a multislice sequence has different imaging parameters. For instance in a MOLLI sequence such as used in this example, it is often the case that each slice has its own set of TI values.
The situation can be accomodated in fit by assigning a
list of dictionaries to the fit_image argument - one for each slice.
We illustrate that here assuming that the TI’s for each slice are offset by
100 ms relative to the previous slice:
molli = [
{
'func': mdreg.fit_abs_exp_recovery_2p,
'TI': TI + 0.1 * z,
}
for z in range(array.shape[2])
]
# Other than that, the function call to `mdreg.fit()` is the same as before.
Other coregistration packages#
This works the same when using ants or elastix for coregistration:
invalid value encountered in cast
The difference with skimage is that the transformations returned are now a 2D array with one transformation at each time and each slice:
print(f'2D transformation shape: {transfo.shape}')
2D transformation shape: (2, 8)
Compare this to 3D registration, where one 3D transformation for each time point is returned:
Elastix coregistration failed. Returning unregistered image. To learn more about the error, set log=True.
3D transformation shape: (8,)
Slice by slice with pixel models#
If the signal model fit is defined with a custom model via the fit_pixels argument, the slice-by-slice operation works the same: set the force_2d keyword to True, and - if each slice has different parameter settings - supply the fit_pixels argument as a list of dictionaries.
Covariance of the parameters could not be estimated
overflow encountered in exp
overflow encountered in multiply
Total running time of the script: (21 minutes 18.343 seconds)